LMU start-up Epicure receives m⁴ Award
7 Jul 2025
The LMU start-up project Epicure has received the m⁴ Award from the Free State of Bavaria for its innovative research into cancer treatment.
7 Jul 2025
The LMU start-up project Epicure has received the m⁴ Award from the Free State of Bavaria for its innovative research into cancer treatment.
The award, which is endowed with 500,000 euros, supports academic research groups in translating promising scientific findings into concrete applications - in this case, the development of novel cancer therapies. Dr. Matthias Heiß, Dr. Corinna Pleintinger, Yasmin Gärtner, Professor Thomas Carell, Dr. Mike Rothe and Professor Franziska Traube from the Department of Chemistry accepted the award in Munich at the beginning of July.
The m⁴ Award is presented every two years by BioM - the network organization of the biotechnology industry in Bavaria - and the Bavarian State Ministry of Economic Affairs, Regional Development and Energy to five outstanding biomedical research projects. The aim is to promote innovative technologies, products or services that contribute to solving urgent medical problems.
Epicure focuses on epigenetically active substances that specifically target disease-relevant molecular mechanisms - with the aim of making cancer treatments more effective and at the same time more tolerable. There is a great need for new therapies, particularly in the area of malignant diseases of the blood and lymphatic system: Despite medical advances, more than 750,000 people worldwide die from hematologic neoplasms every year, as existing treatments are often insufficiently effective or highly toxic.
Thanks to the funding from the m⁴ Award, Epicure can now carry out important preclinical studies that will create the regulatory basis for the first clinical trials in humans - and bring the project one step closer to the planned spin-off.
23 May 2025
LMU has reached a hugely significant milestone in the Excellence Strategy of the German federal and state governments: All seven proposed Clusters of Excellence have successfully passed the review process and will be funded for seven years as of 2026.
The Bavarian Minister of Science, Markus Blume, commented: “Gigantic excellence success for our Munich universities: With seven applications each, including six joint ones, TUM and LMU are successful in the race for the Clusters of Excellence. One thing is clear: Munich is the Mecca of excellence in Germany and sets standards for innovation in Europe. TUM and LMU are the best universities in Germany, and they are proving this once again here. Munich shines today.”
The Faculty of Chemistry and Pharmacy is involved in two of the seven clusters of excellence. The LMU clusters with spokespersons at our Faculty of Chemistry and Pharmacy are:
Understanding and making targeted use of nucleic acids: The NUCLEATE Cluster of Excellence at LMU, the Technical University of Munich (TUM) and Julius-Maximilians-Universität Würzburg (JMU) aims to become a driver of innovation in Germany and Europe by researching the functions and properties of DNA and RNA structures and expanding their immense potential for medical and technological applications.
To this end, the network is highly interdisciplinary and brings together almost all natural science disciplines from organic chemistry and biochemistry, cell and microbiology to medicine and veterinary medicine - supplemented by contributions from computer science and artificial intelligence. NUCLEATE combines outstanding basic research, state-of-the-art technology and application-oriented research. For further translational development, NUCLEATE will work hand in hand with the CNATM future cluster funded by the Federal Ministry of Education and Research - for example in the development of RNA-based therapeutics.
For more information, see: Cluster for Nucleic Acid Sciences and Technologies
Basic research for the energy transition.
How can energy be converted more sustainably and efficiently? The cluster is an innovation platform where researchers are looking for new solutions for photovoltaics, catalysis and batteries.
The joint Cluster of Excellence of LMU and TUM “e-conversion”, which is now entering its second round, is researching fundamental issues of energy conversion in order to find innovative solutions for future applications. The researchers are looking for new approaches for photovoltaics, catalysis and batteries, among other things, with which the global energy demand can be covered more sustainably, efficiently and diversified in the future. As an innovation platform for basic research, the cluster brings together different areas of expertise: from nanoscience and quantum research to semiconductor physics, materials science, computational science and artificial intelligence.
For more information, see: Cluster e-conversion
23 Apr 2025
Philip Tinnefeld from LMU is coordinating a new doctoral training network at the interface of physics, chemistry, biology, and engineering.
The European Union is funding the BioHYBRITE doctoral program to the tune of around 4.2 million euros as part of its Marie Skłodowska-Curie Actions. Entitled “Decoding and designing biomolecular systems with hybrid DNA:RNA:protein nanotechnology,” the program is dedicated to researching novel biomolecular systems.
No fewer than 13 leading academic institutions are collaborating in the Europe-wide network for doctoral training – including LMU Munich, the Technical University of Munich (TUM), the University of Cambridge, the French research center CNRS, and the Max Planck Institute of Biochemistry. The consortium also features several industrial partners, including innovative biotech start-ups. Collectively, they are training 15 doctoral candidates.
Our goal is to train a new generation of highly qualified scientists at the interface of physics, chemistry, biology, and engineering – in the highly promising field of DNA-RNA-protein nanotechnology.Philip Tinnefeld
“BioHYBRITE is a structured, highly interdisciplinary training program at the interface of molecular design, single-molecule analysis, AI-assisted protein development, and medical applications,” says Philip Tinnefeld, Chair Professor of Physical Chemistry at LMU and coordinator of BioHYBRITE. “Our goal is to train a new generation of highly qualified scientists at the interface of physics, chemistry, biology, and engineering – in the highly promising field of DNA-RNA-protein nanotechnology.”
Researchers in this area are working on the development of hybrid nanoscale systems of DNA, RNA, and proteins that can recognize and process information and trigger targeted reactions – similar to living cells.
As for the details, the doctoral researchers in BioHYBRITE receive, among other things, a solid grounding in molecular self-organization and single-molecule analytics. They learn how to design and simulate functional biomolecular systems and use them for applications such as biosensors and targeted drug delivery. This includes training in how to exploit AI for these purposes.
Their training encompasses international summer schools, workshops, laboratory courses, and residencies and internships at academic and industrial partner organizations. Highlights of the program include:
The program is accompanied by regular networking events, a journal club, seminars at the home universities, and personal mentoring sessions. The organizers place particular emphasis on independent working, creative thinking, and international cooperation.
As Tinnefeld explains: “Through the combination of excellent research, methodological breadth, and targeted training in cross-disciplinary skills, BioHYBRITE opens up outstanding career prospects – both in academic research and in the burgeoning bio-nano industry.” The goal is not only to impart state-of-the-art technical knowledge to doctoral candidates, but to prepare them for important roles in science, technology, and society.
9 Apr 2025
LMU researchers have developed a novel cofactor that broadens the use of methyltransferases – with great potential for research and applications.

Chemicist Andrea Rentmeister © Andres Chuquisengo / LMU
Enzymes are highly specific tools of nature which allow chemical reactions to take place under mild conditions and with impressive precision. Transferases are a class of enzymes that transfer carbon chains, so-called alkyl groups, to target molecules. Before now, two major transferase types – methyltransferases and prenyltransferases – have been considered separate enzyme classes, as they transfer alkyl groups of different lengths (with one and five carbon atoms respectively, C1 and C5) and utilize different cofactors.
A team led by Professor Andrea Rentmeister from the Department of Chemistry at LMU has developed a hybrid cofactor that combines features of both systems (S-Adenosyl-L-methionine (SAM) and dimethylallyl diphosphate (DMAPP)). As the researchers report in the journal Chem, the new hybrid cofactor thus permits a broader range of applications. The chimeric compound is accepted surprisingly well by a wide variety of methyltransferases and predominantly leads to so-called prenylation – that is to say, the transfer of larger alkyl groups.
Regarding the background, Rentmeister explains: “Alkylation is an important strategy for influencing chemical and biological properties of small molecules, lipids, nucleic acids, and proteins. Strategies for selective alkylation are therefore extremely important for research and applications.”
Although it is possible to chemically alkylate biomolecules – using compounds such as alkyl halogenides – such reactions are not very selective, which is problematic in the case of complex molecules. By contrast, alkylating enzymes – the biological route, as it were – have proven to be highly selective.
When researchers have wanted to transfer prenyl groups to biomolecules using biotechnological means, the only option available to them has been prenyltransferases. The current research by Rentmeister’s team creates new possibilities. First, the scientists built a chimeric cofactor. “We managed to demonstrate that a cofactor chimera of DMAPP and AdoMet makes methyltransferases act as prenyl-transferring enzymes,” says Rentmeister. The researchers exploited the circumstance that the basic chemical reaction is the same independently of the length of the alkyl residue to be transferred and differs only by dint of the cofactor of the enzymes.
Methyltransferases, which ordinarily transfer methyl groups to molecules, did likewise with prenyl groups with the same specificity in the presence of the chimeric cofactor AdoPrenyl. Rentmeister’s team showed that with modified cofactors it is possible to connect, for example, chains containing 10 or 15 carbon atoms to various biomolecules
Most notably: “The demonstration that naturally occurring methyltransferases can transfer prenyl groups contradicts the prevailing textbook opinion of a strict separation between C1- and C5-transferring enzyme classes,” says the scientist. This has delivered a blow to a fundamental dogma of enzyme chemistry – and opened up new questions about the underlying reaction mechanisms.
ould have promising applications in the field of protein chemistry. The targeted modification of proteins with longer-chained alkyl groups can change their biological activity and their localization in the cell or influence their recognition by other molecules. This makes alkylation an important tool in biotechnology, medicine, and pharmacy.
Nicolas V. Cornelissen, Arne Hoffmann, Pulak Ghosh, Yanis L. Pignot, Mehmet Erguven, Andrea Rentmeister: Chimeric cofactors enable methyltransferase-catalyzed prenylation. Chem, 2025
20 Mar 2025
LMU scientist Olivia Merkel researches nanocarriers for the targeted delivery of drugs to their site of action. Now the company she co-founded, RNhale, has been awarded a lucrative EU grant to bring a new anti-asthma therapy to clinical readiness.

Investigates novel nanocarrier systems: Olivia Merkel. | © LMU
The company name describes the envisioned product in a nutshell: RNhale – that is, RNA therapeutics to inhale. Now the young Munich-based company has been awarded a Transition grant of 2.5 million euros by the European Innovation Council (EIC) to bring such a novel anti-asthma drug to clinical practice.
The company RNhale is an LMU spin-off: Olivia Merkel, Chair Professor of Drug Delivery, and her team have built up the requisite know-how over many years. The pharmaceutical researcher investigates novel nanocarrier systems capable of delivering drugs to specific sites of action in the human body. Her main research focus is on the therapeutic application of short sections of RNA that can silence genes involved in pathogenesis in certain cell types.
On this basis, Merkel and her team developed approaches for new anti-asthma therapies, with funding from sources such as a multi-million euro Starting Grant from the European Research Council (ERC). The researchers packed specific so-called siRNA into nanocarriers that can be stabilized by means of spray drying and processed into an inhalable dry powder.
To enable Merkel to put such therapeutics to the test, she was awarded a Proof of Concept Grant by the ERC. Following the receipt of additional funding from LMU’s Knowledge Transfer Fund, the conditions were right to found the spin-off RNhale, for which Merkel acts as scientific advisor. The founder team includes various colleagues at LMU, including the CEO Benjamin Winkeljann.

CEO Benjamin Winkeljann is part of the founder team. | © Joshua Winkeljann
The new EIC grant is the logical next link in this funding chain from Brussels. RNhale will use the grant to prepare the technology for clinical studies and drive forward the development of the business for market release. This includes preparing the requisite preclinical trials.
As an initial application, the company plans to develop a drug to reduce expression of the cytokine TSLP in the respiratory tract, which occurs in patients with allergic asthma. Through this work, RNhale ultimately hopes not only to bring a highly effective anti-asthma therapeutic to market, but also to create a platform that can be used to develop drugs to treat other respiratory conditions.
As a University of Excellence, LMU supports its scientists in translating the huge innovation potential of basic research into specific fields of application. To this end, LMU provides its own researchers with a comprehensive range of advice services. More about research transfer services at LMU
20 Mar 2025
Chemists reveal method for differentiating PCET mechanisms – a key step for steering fundamental energy conversion and redox catalysis processes.
Redox reactions form the basis of many fundamental processes of life. Without them, neither cellular respiration nor photosynthesis could take place. Redox reactions also play a crucial role in applications in the domains of chemistry, biochemistry, and the use of light for energy generation. Understanding the fundamental principles of these reactions is therefore important for driving forward new technologies. Using an innovative method based on high pressures, a team led by LMU chemist Professor Ivana Ivanović-Burmazović, member of the “e-conversion” Cluster of Excellence, and Professor Dirk Guldi from FAU Erlangen-Nürnberg has managed for the first time to differentiate two related reaction mechanisms.
In redox reactions, electrons are transferred between molecules. Because electrons have a negative charge, this can cause the charge of the reactants to change, which is energetically demanding. Nature has found an elegant solution to prevent this: Often, the transfer of electrons is coupled with the transfer of positively charged protons. This proton-coupled electron transfer (PCET), as it is known, does not produce any change in charge – the most efficient way for a redox reaction to occur.
There are two possible mechanisms here: Either electrons and protons are transferred simultaneously (“concerted”), or the transfer occurs in stepwise fashion – that is to say, with electrons and protons transferred separately. “To be able to optimize these processes, we need to know the exact mechanisms,” says Ivanović-Burmazović. “Before now, however, there has been no direct method for differentiating the two alternatives with certainty. Our work set out to remedy this.”
For their study, the researchers investigated the influence of pressure on the very rapid (within nanoseconds) light-induced reaction of a photosensitive molecule in solution. It was already known that this molecule transfers both protons and electrons to corresponding acceptor molecules, but the exact course of these processes – the mechanism – was unknown. “Our results show that measuring the effect of pressure on the reaction rate allows direct inferences to be drawn about the mechanisms,” explains Ivanović-Burmazović.
If high pressure – in the experiment, up to 1,200 atmospheres – is applied and the reaction rate remains unchanged, it is a concerted reaction. “When electrons and protons are transferred simultaneously, charge of reacting species does not change and neither does the associated solvation sphere – that is, the cluster of solvent molecules surrounding the molecules. Therefore, pressure has no influence on reaction rate – a clear sign of a concerted mechanism,” explains Ivanović-Burmazović. If the rate changes, however, this points to changes in the charge and to a change in the volume of the solvation sphere – indicating a stepwise process.
To their surprise, the researchers were able not only to determine the type of mechanism, but also influence the process: “By increasing the pressure, we managed to steer the reaction from a stepwise mechanism toward a concerted mechanism,” says Ivanović-Burmazović.
The new findings are highly significant for numerous research areas that deal with the motion of electrons and protons, emphasize the authors. They not only offer new insights into fundamental chemical processes, but could also help advance new technologies concerned with the conversion and storage of chemical energy – such as redox catalysis for the generation of solar fuels or for hydrogen production.
Daniel Langford, Robin Rohr, Stefan Bauroth, Achim Zahl, Alicja Franke, Ivana Ivanović-Burmazović and Dirk M. Guldi: High-pressure pump–probe experiments reveal the mechanism of excited-state proton-coupled electron transfer and a shift from stepwise to concerted pathways. Nature Chemistry 2025
27 Jan 2025
Andrea Rentmeister researches how to visualize and control molecular processes in cells.
“Whereas classical chemistry works with test tubes and round-bottomed flasks, we go a step further: living cells are our test tubes,” explains Professor Andrea Rentmeister, who has been Chair of Organic and Biological Chemistry at LMU’s Faculty of Chemistry and Pharmacy since the start of 2024. Her work operates at the boundaries of chemistry and biology – with the goal of decoding and actively controlling fundamental processes in the cell. “We’re trying to change biomolecules such as mRNA using chemical and biological methods, so that we can better analyze them or improve their properties.”
When you look inside a cell, you do not see much at first. But if you attach synthetic labels to certain biomolecules, you can observe them better under the microscope. With chemical modifications, moreover, you can change the function of biomolecules – by switching their activity on or off, for example, using light-dependent markers.
However, this is easier said than done. “In a cell, there are countless components that can react with each other,” explains the chemist. “If I add a new molecule, I have to consider that some other molecule could react with it – and that’s extremely complex.” The solution is a technique known as click chemistry or bioorthogonal chemistry. This involves developing so-called bioorthogonal groups, which, all going well, react exclusively with each other and with nothing else in the cell. “Bioorthogonal chemistry opens up an entirely new dimension,” says Rentmeister. “You build a molecular lock, so to speak, and a matching key. The two click with each other, but don’t react with anything else.”
Whereas classical chemistry works with test tubes and round-bottomed flasks, we go a step further: living cells are our test tubes.PROF. DR. ANDREA RENTMEISTER
As part of her doctoral thesis, Rentmeister carried out research into RNA in connection with in-vitro evolution – “a fascinating process that involves ‘breeding’ molecules with desirable properties.” Later, in California, she researched under Frances H. Arnold, who received a Nobel Prize in 2018 for her work on the directed evolution of proteins. In her own work, Rentmeister tried to connect the RNA and the protein levels. “Naturally enough, I didn’t want to just emulate my boss, but also do my own thing. My work centered around changing proteins so they can be used to modify RNA.”
Over time, she increasingly incorporated chemical approaches into her research. After her postdoc period in California, Rentmeister was Junior Professor at the University of Hamburg. Then, before her appointment at LMU, she spent several years in the Institute of Biochemistry at the University of Münster, where she was Professor of Biological Chemistry and Biomolecular Label Chemistry.
Andrea Rentmeister sees herself as a basic researcher. Her primary goal is to decode fundamental chemical and biological realities. At the same time, her work has great potential for practical applications – for example, in mRNA-based cancer therapy. Her methods could make it possible one day to selectively obtain effects in specific cells without affecting other cells, which would considerably increase the safety and efficacy of mRNA-based therapies. But there is still a long way to go before such treatments are a reality, cautions Rentmeister. “Currently, we’re working on establishing such principles in model organisms like the zebrafish to test their feasibility.”
When we ask the chemist which topic she is most excited about at the moment, she finds it hard to pick: “All my projects are my favorites!” Currently, her energies are particularly devoted to the cell-selective activation of translation, “a topic we’re addressing as part of a new German Research Foundation (DFG) project.” In addition, she’s doing intensive research in the fields of epigenetics and epitranscriptomics – that is to say, modifications of DNA and RNA that influence the activity and regulation of genes. “Our goal is to take molecules that used to be hard to capture in biology and make them accessible and analyzable.” Again, chemical tools are very much part of her toolkit here.
Our goal is to take molecules that used to be hard to capture in biology and make them accessible and analyzable.PROF. DR. ANDREA RENTMEISTER
Has she ever had something like a eureka moment? Yes, says the chemist, from time to time, although brainwaves do not appear from nowhere: “Chance favors the prepared mind, as Louis Pasteur put it. You work on a topic intensively over a long period and then – usually at a moment when you’re not expecting it – you get an insight. Although the eureka moment feels sudden and unexpected, it has actually matured in the mind over years of research.”
Another vital factor is having the right work environment. The Faculty of Chemistry and Pharmacy at LMU offers ideal conditions for her endeavors. At the adjacent Gene Center Munich and the surrounding institutes, there is a plethora of research groups that focus on nucleic acids. In her specialist area, she explains, the density of excellent researchers with different professional backgrounds is particularly high here. “When chatting to people in the office kitchen, you discover things that are not in print, gain new perspectives and impressions, and sometimes even get exciting new research ideas.”
9 Jan 2025
An interdisciplinary study conducted within the e-conversion Cluster of Excellence demonstrates the huge potential of the crystalline semiconducting structures.

Laura Spies looks at one of the COF thin films examined in the study. | © Florian Wolf
An interdisciplinary research team from LMU, the Technical University of Munich (TUM), and the University of Oxford has employed novel spectroscopic techniques to investigate the diffusion of excited states in so-called covalent organic frameworks (COFs). These modular materials can be adapted for desired properties through the targeted selection of their components, offering a broad range of applications. The study revealed how efficiently energy can be transported in these crystalline, semiconducting materials – a decisive advance for future optoelectronic applications such as sustainable photovoltaic systems and organic light-emitting diodes (OLEDs).
At the heart of the study, which has been published in the renowned Journal of the American Chemical Society, we find COF thin films of highly crystalline, porous material. Through the use of state-of-the-art spatiotemporal techniques like photoluminescence microscopy and terahertz spectroscopy in conjunction with theoretical simulations, the team revealed remarkably high diffusion coefficients and diffusion lengths of several hundreds of nanometers. “As such, these thin films significantly exceed the known energy transport capabilities of similar organic materials,” emphasizes Laura Spies, doctoral candidate at the Chair of Physical Chemistry and Functional Nanomaterials at LMU and co-lead author. “The energy transport works exceptionally well, even across structural defects such as grain boundaries,” adds Dr. Alexander Biewald, former doctoral candidate in the Physical Chemistry and Nanooptics group and second co-lead author of the study.
Temperature analyses yielded further insights into the underlying mechanisms. “The results indicate that both coherent and incoherent transport processes are at play,” explains Professor Frank Ortmann, co-author of the study. Coherence pertains when the waves of motion occur in an orderly fashion, undisturbed over long distances, allowing fast and low-loss energy transfer. Incoherent processes, by contrast, are characterized by disordered, random motions, which require thermal activation and are often less efficient. These insights significantly contribute to our understanding of energy transport in COFs and show how the molecular structure and organization in the crystal can affect these processes.
“Our work highlights how vital the interdisciplinary and international cooperation of researchers with expertise in synthesis, experimental analysis, and theoretical modeling – made possible by e-conversion – is for the success of such studies,” say the corresponding authors of the study, Professor Achim Hartschuh and Professor Thomas Bein. The results open up new prospects for the development of sustainable organic materials in photocatalysis and optoelectronics, such as photovoltaics.
L. Spies et al.: Spatiotemporal Spectroscopy of Fast Excited-State Diffusion in 2D Covalent Organic Framework Thin Films, Journal of the American Chemical Society (JACS) 2025.
13 Dec 2024
Bettina Valeska Lotsch receives the prestigious DFG prize - the highest scientific award in Germany.

Prof. Bettina Lotsch
With the award of the prestigious Gottfried Wilhelm Leibniz Prize, the DFG is recognizing the outstanding research achievements of our honorary professor Bettina Valeska Lotsch.
Bettina Valeska Lotsch is being honored for her work in solid-state chemistry between basic materials synthesis and the development of new materials.
The Gottfried Wilhelm Leibniz Prize of the German Research Foundation is considered the most important award in German science. This year, ten scientists will receive it. The prize money of 2.5 million euros can be used by the recipients for their scientific work for up to seven years.
5 Dec 2024
On November 29, Römer prizes were awarded and diplomas were presented to the graduates of the Master's degree courses in Chemistry and Biochemistry.

Foto: Michael Till / Gene Center LMU | © (c) LMU, till@genzentrum.lmu.de
On November 29, the Römer Prizes for outstanding achievements by young scientists from the Departments of Biochemistry and Chemistry were awarded for the 19th time. As part of a large ceremony, first, the diplomas were presented to the graduates of the Master's degree programmes in Chemistry and Biochemistry and then the Römer Prizes were awarded.
The following received Römer prizes totalling € 500 for outstanding achievements in their Master's thesis:
In addition, the following were honoured with Römer Prizes of € 1,250 for their Phd thesis:
Finally, Prof. Dr. Dr. h.c. Peter H. Seeberger from the Max Planck Institute of Colloids and Interfaces gave this year's keynote speech ‘Vaccines from sugar and drugs from plant waste’.
27 Nov 2024
LMU researchers have investigated how cationic polymers organize on a molecular level when transporting RNA drugs.

Olivia Merkel researches nano transport systems for the targeted administration of drugs. | © Florian Generotzky
Cationic polymers are promising tools for transporting RNA therapeutics or RNA vaccines. Like lipid nanocarriers, they are used to deliver mRNA medicines. The nanoscopic packaging materials are able to effectively protect their load and deliver them to the target cells. “We manufacture so-called ‘gene ferries,’ into which all kinds of therapeutic nucleic acids can be encapsulated for secure transport to the site of action,” explains Professor Olivia Merkel, Chair of Drug Delivery at LMU’s Faculty of Chemistry and Pharmacy.
To further improve the effectiveness of these gene ferries, however, it is important to understand how these particles organize on a molecular level, encapsulate RNA, and release it again – an aspect which so far has not been fully examined. Merkel is principal investigator of a new study that has yielded fresh insights into the organization of the nanocarriers. The study was carried out as part of her ERC research project RatInhalRNA (Rational and Simulation-Supported Design of Inhalable RNA Nanocarriers) and the findings were recently published in the journal Nano Letters.
We manufacture so-called ‘gene ferries,’ into which all kinds of therapeutic nucleic acids can be encapsulated for secure transport to the site of action.Olivia Merkel
“Our research used a technique called coarse-grained molecular dynamics (CG-MD) to simulate and visualize the particles,” explains Merkel. The specific focus was on how changes in polymer structure and environmental conditions impact particle formation. Simulations were supported by wet lab experiments using nuclear magnetic resonance (NMR), which confirmed that CG-MD can reveal detailed insights into the structure and behavior of RNA nanoparticles. “This study highlights CG-MD’s value in predicting and explaining the properties of RNA nano-formulations, which can help in designing better systems for future medical applications,” says Merkel.
Katharina M. Steinegger, Lars Allmendinger, Sebastian Sturm, Felix Sieber-Schäfer, Adrian Philipp Eckart Kromer, Knut Müller-Caspary, Benjamin Winkeljann & Olivia M. Merkel: Molecular Dynamics Simulations Elucidate the Molecular Organization of Poly(beta-amino ester) Based Polyplexes for siRNA 3 Delivery. Nano Letters 2024
22 Nov 2024
LMU researchers have discovered how the interplay between a key protein and an endolysosomal ion channel promotes tumor development in skin cancer.
Melanoma arising from pigment-producing cells known as melanocytes is the deadliest form of skin cancer. A major cause of melanoma is excessive exposure to ultraviolet light, from sunlight or other sources, which can trigger mutations that promote tumor formation. A team led by LMU pharmacologist Professor Christian Grimm (Walther Straub Institute of Pharmacology and Toxicology) and Dr. Karin Bartel (Faculty of Chemistry and Pharmacy) has now investigated the molecular mechanisms of tumorigenesis. As the researchers demonstrate, the interplay of two proteins – the ion channel TPC2 and the enzyme Rab7a – plays a decisive role, as they promote the growth and metastasis of melanoma.
Studies have shown that certain activity-boosting mutations in the ion channel TPC2 are associated with fair skin, blond hair, and albinism. These traits make people particularly susceptible to melanoma, as their skin offers less protection against harmful ultraviolet radiation. Conversely, loss of TPC2 is associated with decreased melanoma risk. The ion channel controls the breakdown of important proteins in endolysosomes – cell organelles that are involved in transport and degradation processes – and thus influences signaling pathways that regulate tumor growth.
Like TPC2, the protein Rab7a is an important regulator for the endolysosomal system. Earlier proteome analyses had shown, moreover, that Rab7a is a potential interaction partner of TPC2. Using modern methods such as endolysosomal patch-clamp electrophysiology and the measurement of lysosomal calcium release via fluorescence microscopy, the researchers established that there was indeed an interaction between Rab7a and TPC2 at the functional level, which promoted the growth and invasiveness of melanoma cells. Conversely, the pharmacological inhibition of Rab7a decreased TPC2 activity and thus melanoma growth.
“Our results show that Rab7a, by amplifying TPC2 activity, plays a key role in the regulation of tumor growth,” says Grimm. “Specifically, the activation of TPC2 by Rab7a reduces the levels of a certain protein. This protein boosts the stability of a transcription factor that is a key regulator in melanocytes and melanomas and promotes their proliferation and survival.”
The interaction between Rab7a and TPC2 could pave the way for new therapeutic strategiesChristian Grimm
A particularly notable finding, according to the researchers, was that effects of the interaction of Rab7a and TPC2 could be demonstrated in vivo. In mouse models with melanoma cells without Rab7a or TPC2, they found that tumor size and metastasis were much reduced. “The interaction between Rab7a and TPC2 could pave the way for new therapeutic strategies which target the specific signaling pathways that promote melanoma growth and metastasis” concludes Grimm.
C. Abrahamian et al.: Rab7a is an enhancer of TPC2 activity regulating melanoma progression through modulation of the GSK3β/β-Catenin/MITF-axis. Nature Communications 2024
8 Nov 2024
LMU researchers have developed a new method for capturing structural changes and interactions of DNA and proteins at high resolution.
For fundamental processes of life such as the replication, transcription, and repair of DNA, the complex interplay between DNA and proteins plays a decisive role. A team led by LMU chemist Professor Philip Tinnefeld has developed a new fluorescence microscopy method that is capable of visualizing dynamic changes in the structure of DNA and its interaction with proteins at a spatial resolution down to the angstrom range and with a temporal resolution of under a second.
To this end, the researchers exploited a combination of two effects. Firstly, they were able to measure the distance of a dye molecule to a graphene surface, because the dye appears darker – or has a shorter fluorescence lifetime – the closer it is to the graphene. Secondly, they managed to immobilize the DNA on the graphene in such a way that the DNA strands were arranged vertically.
“This allowed us to know the spatial orientation of the DNA during measurements,” explains Tinnefeld. From the changes in brightness of the dye, the researchers can then identify structural changes in the DNA or the movements of proteins along the DNA. “This can decisively advance the analysis of fundamental processes such as DNA repair mechanisms and other important applications,” says Tinnefeld. “Our method has the potential to develop into a widely used technology and open up new opportunities in structural biology and for biosensor systems and related 2D materials.”
A.M. Szalai, G. Ferrari, L. Richter et. al: Single-Molecule Dynamic Structural Biology with Vertically Arranged DNA on a Fluorescence Microscope. Nature Methods 2024
7 Nov 2024
LMU researchers have developed a strategy that enables biosensors to be easily adapted for a wide range of applications.

Dr. Viktorija Glembockyte | © LMU
Biosensors play a key role in medical research and diagnostics. At present, however, they generally have to be specially developed for each application. A team led by LMU chemist Philip Tinnefeld has developed a general, modular strategy for designing sensors that can be easily adapted to various target molecules and concentration ranges. As the researchers report in the journal Nature Nanotechnology, their new modular sensor has the potential to significantly accelerate the development of new diagnostic tools for research.
The sensor uses a DNA origami scaffold, which consists of two arms connected by a molecular “hinge.” Each arm is tagged with a fluorescent dye, and the distance between the tags is recorded by means of fluorescence resonance energy transfer (FRET). In a closed state, the two arms are parallel; when the structure opens, the arms fold out to form an angle of up to 90°. “As a result of this large conformational change, the fluorescence signal also changes substantially,” explains Viktorija Glembockyte, senior author of the study. “This allows signals to be measured with considerably greater clarity and precision than in systems with small conformational changes.”
The origami scaffold can be equipped with docking sites for various biomolecular targets such as nucleic acids, antibodies, and proteins. Whether the sensor is open or closed depends on the binding of the respective target molecule to the origami scaffold. The sensor can thus be deliberately adapted and optimized through the use of additional binding sites or stabilizing DNA strands. “It’s relatively easy to design the origami such that several molecular interactions between target molecule and sensor are queried simultaneously,” explains Tinnefeld. “These multiple bonds lead to interesting cooperative effects which make it possible to specifically control the sensitivity of the sensor without intervening in the biomolecular interactions themselves – that is to say, the strength with which the target molecule docks to its binding site. This flexibility is a major advantage of our system.”
The researchers plan to further optimize the sensor in the future for biomedical and other applications. A possible field of application could be sensors that monitor various parameters and release active agents under certain conditions, says Tinnefeld
L. Grabenhorst et al.: Engineering Modular and Tunable Single Molecule Sensors by Decoupling Sensing from Signal Output. Nature Nanotechnology 2024
29 Oct 2024
Live lecture by Prof. Ivana Ivanović-Burmazović at the Munich Science Days
What do ageing processes and inflammation in our bodies have in common with processes in a fuel cell and the principles of chemical and biological energy conversion?
Find out for yourself in this lecture by our Director of Inorganic Chemistry.
26:19 | 29 Oct 2024 | ©Münchner Wissenschaftstage
21 Oct 2024
In Professor Silvija Markic's LMUchemlab, pupils can experience chemistry in an inclusive environment on their own terms.
Professor Silvija Markic conducts research into linguistic heterogeneity and cultural diversity in chemistry lessons as well as the learning and teaching of technical languages at school and university. Together with her doctoral students, such as Jannis Memmen, she supervises the LMUchemlab, the Chemistry Department's learning laboratory. Here, students can experiment independently in a supervised environment - without time pressure and adapted to their individual learning needs.
The LMUchemlab has the concept that we want to reach all students. Regardless of their language skills, cultural background or socio-economic status.Silvija Markic, Professor for the didactics of chemistry
2:42 | 24 Oct 2024
18 Sept 2024
LMU chemists present two studies that open up new possibilities for biotechnological applications.
In the world of nanotechnology, the development of dynamic systems that respond to molecular signals is becoming increasingly important. The DNA origami technique, whereby DNA is programmed so as to produce functional nanostructures, plays a key role in these endeavors. Teams led by LMU chemist Philip Tinnefeld have now published two studies showing how DNA origami and fluorescent probes can be used to release molecular cargo in a targeted manner.
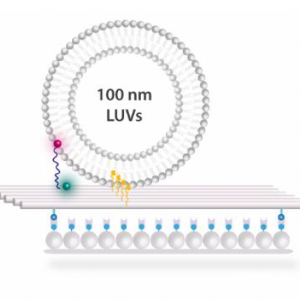
In the journal Angewandte Chemie, the researchers report on their development of a novel DNA-origami-based sensor that can detect lipid vesicles and deliver molecular cargo to them with precision. The sensor works using single-molecule Fluorescence Resonance Energy Transfer (smFRET), which involves measuring the distance between two fluorescent molecules. The system consists of a DNA origami structure, out of which a single-stranded DNA protrudes, which has been labeled with fluorescent dye at its tip. If the DNA comes into contact with vesicles, its conformation changes. This alters the fluorescent signal, because the distance between the fluorescent label and a second fluorescent molecule on the origami structure changes. This method allows vesicles to be detected.
In a second step, the system can be used as a means of transporting molecules, with the sensing strand serving as molecular cargo that can be transferred to the vesicle. Through a further modification of the system, the researchers were also able to precisely control the transfer of the cargo.
Lipid vesicles play a key role in many cellular processes, such as molecular transport and signal transmission. As such, the ability to detect and manipulate them is particularly interesting for biotechnological applications like the development of targeted therapies. The approach laid out here could show a way to load lipid nanoparticles with a precisely defined number of molecules in applications such as vaccines. “Our system also offers promising approaches for biological research when it comes to better understanding and controlling cellular processes at the molecular level,” says Tinnefeld.
In the second study, which was recently published in the journal Nature Communications, a second team led by Tinnefeld and Yonggang Ke (Emory University, Atlanta, Georgia) presents a DNA origami structure which undergoes a stepwise allosteric conformational change when certain DNA strands bind. Using FRET probes, the researchers were able to track this process at the molecular level and show how the reaction steps can be temporally controlled. In addition, they demonstrate how a DNA cargo can be released in a targeted manner during this process, opening up new opportunities for controlled reaction cascades.
E. Büber et al.: DNA Origami Vesicle Sensors with Triggered Single-Molecule Cargo Transfer. Angewandte Chemie 2024
F. Cole et al.: Controlled mechanochemical coupling of anti-junctions in DNA origami arrays. Nature Communications 2024
12 Jul 2024
The research group of LMU pharmacist Olivia Merkel has optimized the synthesis of polymer nanoparticles to facilitate the targeted distribution of RNA agents in the body.

Olivia Merkel is the lead author of the newly published study. | © Florian Generotzky / LMU
RNA therapy with polymer nanoparticles is considered a promising approach for the treatment of various illnesses. It involves the use of polymers as “nanocarriers” to transport RNA drugs precisely to the correct target cells. Manufacturing such polymers, however, has proven to be complex and difficult.
In a recent study by the research group of Olivia Merkel, Professor of Drug Delivery in the Department of Pharmacy at LMU, so-called spermine-modified poly(beta-amino esters) (PBAEs), a polymer type that is frequently used for the formulation and delivery of nucleic acids were focused on. “We synthesized and characterized a library of 27 different polymers, taking into account various factors such as the ratios of starting materials, the temperature, and the reaction time,” explains Merkel.
Our research helps to improve the quality, efficiency, and precision of RNA drugsOLIVIA MERKEL

Factors that significantly influence polymerization are identified by statistical analysis of a library of 27 polymers that are particularly suitable for therapeutic RNA delivery. | © Merkel Lab
A design-of-experiment approach, whereby the significant factors of experiments are identified by means of statistical analyses, enabled the researchers to derive a wealth of information out of just a few experiments. The polymers were chemically analyzed to understand their composition and molecular properties. In addition, a computer-based protocol was developed to better capture the complex process of polymerization and predict it for future syntheses. “Our research helps to improve the quality, efficiency, and precision of RNA drugs,” says Merkel regarding the results of the study.
Adrian Kromer, Felix Sieber-Schäfer, Johan Farfan Benito & Olivia M. Merkel: Design of Experiments Grants Mechanistic Insights into the Synthesis of Spermine-Containing PBAE Copolymers. ACS Publications 2024
12 Jul 2024
On the ‘Day for Good Teaching’ at LMU on 12 July, LMU research prizes for excellent students and LMU teaching innovation prizes for lecturers will be awarded.
Two students from the Faculty of Chemistry and Pharmacy were honoured for their work:
„Ni(II)-Catalysed Negishi Cross-Coupling Reactions with Chiral Pyridine-Hydrazone Ligands for Atroposelective Biaryl Synthesis“
Faculty of Chemistry and Pharmacy
Damian Groß
Damian Groß has developed and optimised a new method for the stereoselective synthesis of biaryl atropisomers. These compounds are of great importance as reagents, catalysers and medicines and form important structures for new functional molecules in technology and medicine. Groß applied a wide range of preparative organic chemistry laboratory techniques as part of this work. The results he achieved will enable a broad application of this methodology in the future, in particular for the synthesis of new ligands and other natural substances as well as drugs. The easy accessibility of the catalyst system, its simple realisation and the fact that it does not require expensive reagents are particularly noteworthy. His results have already been published in a renowned scientific journal.
„Design and synthesis towards Trx-selective inflammatory-regulating prodrugs utilising self-immolative spacers”
Faculty of Chemistry and Pharmacy
Julia Brandmeier
Julia Brandmeier's project enabled the design, synthesis and already practical applications of a new class of diagnostics for understanding metabolism and therapeutic drug candidates. These were developed to interact with one of the most important metabolic networks in cells, the thioredoxin system of thiol/disulphide oxidoreductases. It plays an essential role in DNA synthesis and inflammatory reactions and regulates the repair of damage in cells. Until now, it has been difficult to selectively target this enzyme class with small molecules. By overcoming the barrier in the adaptation between certain disulphide enzymes and amine-containing agents, the work has a strong model character. Brandmeier's research encompassed fundamental chemistry and organic synthesis, chemical biology, pharmaceutical and medicinal chemistry, biochemistry and cell biology. It is already being transferred to applications at LMU and developed further - from diagnostics to antiproliferative cancer therapeutics.
17 Jul 2024
On 7 June, our faculty once again hosted the popular university day for pupils.

On the University Day, a group of selected high school students once again visited our faculty to find out more about studying chemistry and biochemistry. The program included a scientific lecture on catalysis, a visit to a regular biochemistry lecture and the opportunity to conduct their own experiments in the research laboratories of the Chemistry and Biochemistry Departments.
16 Jul 2024
At its annual meeting in Italy, the Controlled Release Society (CRS) presented two awards to Prof Olivia Merkel for her services to the society.

Foto: Credits CRS
The „Controlled Release Society“ (CRS) held their Annual Meeting in Bologna, Italy, from July 8-12, and Prof. Olivia Merkel received not only one but two awards for her service to the society.
She was inducted into the College of Fellows of the CRS, and she received the Skin and Mucosal Delivery Focus Group’s “Medal of Honor”. Olivia Merkel was the president of the German Chapter of the CRS in 2020 and the Chair of the Skin and Mucosal Delivery Focus Group from 2020-2022. It was a busy week as she also participated in a panel discussion together with ERC President Prof. Maria Leptin and other ERC awardees, presented her research in an invited talk and participated in two editorial board meetings. The CRS is home for experts dedicated to drug delivery science.
16 Jul 2024
The European Research Council has awarded the LMU chemist a grant for the preclinical development of a new drug for treating leukemia in high-risk patients.

Professor Thomas Carell. | © LMU
Professor Thomas Carell, Head of the Institute of Chemical Epigenetics at LMU, has been funded by the European Research Council (ERC) since 2016 with a highly endowed ERC Advanced Grant. The chemist has now been awarded a Proof of Concept Grant to build on this. With this program, the ERC supports researchers in transferring their results from research into practice.
Thomas Carell is researching how and why the organism chemically modifies nucleic acids such as the genetic molecules DNA and RNA. One such modification is the insertion of methyl groups by enzymes known as methyltransferases. Methylation is important for the regulation of gene activity, but it can also be involved in the development of cancer. So-called hypomethylating agents specifically inhibit methyltransferases and are regularly used to treat diseases of the hematopoietic system such as AML (acute myeloid leukemia) and MDS (myelodysplastic syndrome) in older patients with poor general health.
However, treatment options are currently limited due to the instability of these active substances. Despite research into alternative therapeutic approaches, there is therefore a considerable need for mild treatment options with high success rates for this patient group. This is where Carell comes in. With the active substance carbacitabine (CAB), he was able to develop a more stable active substance that showed better efficacy and lower toxicity than previous therapeutic agents in the AML mouse model. CAB is therefore a promising candidate for the treatment of leukemia in high-risk patients.
In his new project leuCAB (Preclinical Development of new nucleoside-based drug against leukemia), Carell now wants to drive forward key preclinical steps in order to exploit the potential of CAB. This includes further optimization of the structure of the active substance and comprehensive preclinical studies to evaluate the safety and efficacy of CAB. In addition, CAB synthesis is to be increased in order to meet the demand for preclinical and clinical studies. In order to conduct clinical trials, Carell intends to drive forward the establishment of a spin-off company and involve various key stakeholders. "This comprehensive approach will ensure the effective translation of CAB from the laboratory to the clinic and give new hope to patients, especially those struggling with AML and MDS," says Carell.
10 Jul 2024
Regina de Vivie-Riedle, born in Wuppertal in 1958, passed away after a long, serious illness on June 20, 2024 at the age of 66.

After studying chemistry at the Friedrich Wilhelm University of Bonn, Regina de Vivie-Riedle received her doctorate from the University of Bonn in 1987 under Professor Sigrid D. Peyerimhoff. This was followed by a post-doctorate at the Max Planck Institute for Quantum Optics in Garching (1988 - 1990) and at the University of Colorado, Boulder, USA (1991 - 1992). Supported by a habilitation scholarship from the German Research Foundation, she worked as a research assistant at the Institute for Physical and Theoretical Chemistry at the Free University of Berlin from 1994 to 1997, where Regina de Vivie-Riedle completed her habilitation in theoretical chemistry in 1997.
After a position as C3 group leader at the Max Planck Institute for Quantum Optics, Garching (1997-2002), Regina de Vivie-Riedle was an adjunct professor of theoretical chemistry at the Faculty of Chemistry and Pharmacy at the Ludwig Maximilian University of Munich from 2002 until the end of her life.
De Vivie-Riedle's research focused on the numerical description of chemical dynamics using quantum dynamic methods. Early on, de Vivie-Riedle was involved in pioneering topics such as the quantum mechanical control of chemical reactions using light fields and quantum computing based on molecular systems. Her research has provided profound microscopic insights into the dynamics of chemical reactions. During her career, de Vivie-Riedle has been instrumental in shaping the field of theoretical femtochemistry in Germany.
Regina de Vivie-Riedle not only made outstanding scientific achievements in the field of theoretical chemistry and quantum dynamics, but also provided outstanding support to the faculty over many years as Dean of Studies and Women's Representative. Her legacy will endure through the numerous publications and the generations of chemists she has trained and inspired. In Regina de Vivie-Riedle we have lost a great, highly esteemed colleague who inspired us both scientifically and personally. We will sorely miss her.
25 Jun 2024
LMU chemists have successfully synthesized Ruddlesden-Popper nitrides for the first time, opening the door to new materials with unique properties.
Ruddlesden-Popper compounds are a class of materials with a special layered structure that makes them interesting for numerous applications – as superconductors or catalysts, for example, or for use in photovoltaics. There have been many halides and oxides of this structural type before now, but no nitrides. Although scientists expected Ruddlesden-Popper nitrides to have outstanding material properties, they were unable to actually manufacture them.
Now researchers led by Dr. Simon Kloß from the Department of Chemistry at LMU have developed a special synthetic pathway which has enabled them to manufacture nitride materials that crystallize in the Ruddlesden-Popper structural type, as they report in the journal Nature Chemistry.
The stability of the triple bond in the nitrogen molecule (N2) and the low electron affinity of the element made it very challenging for the chemists to manufacture the nitrogen-rich Ruddlesden-Popper nitrides. They achieved a breakthrough by carrying out the syntheses under extreme conditions. Employing large-volume presses, they compressed their samples at pressures of eight gigapascals, which is equivalent to 80,000 bars. Then they used an active nitrogen source such as sodium azide to prepare the rare-earth transition-metal nitride compounds.
“We think we can systematically investigate Ruddlesden-Popper nitrides compounds with our new synthesis strategy,” says Kloß. The scientists demonstrated this by investigating three new compounds of this materials class – a cerium-tantalum nitride (Ce2TaN4) and praseodymium- and neodymium-rhenium nitrides (Ln2ReN4 (Ln = Pr, Nd)). “These three initial materials already exhibit a rich variety of structural, electronic, and magnetic properties,” says Kloß.
The praseodymium and the neodymium compounds displayed exciting magnetic characteristics. For example, the neodymium compound is a remarkable hard ferromagnet with irreversible magnetic behavior. Meanwhile, the tantalum compound is a semiconductor with properties that make it exciting for applications in the energy conversion domain or as a ferroelectric material. “The same synthetic method will probably lead to other Ruddlesden-Popper nitride compounds and their derivatives,” explains Kloß. “Consequently, a large new class of nitrides is waiting to be researched.”
M. Weidemann, D. Werhahn, C. Mayer, S. Kläger, C. Ritter, P. Manuel, J. P. Attfield & Simon D. Kloß: High-pressure synthesis of Ruddlesden–Popper nitrides. Nature Chemistry (2024)
17 Jun 2024
Eyes glued to a live transmission from inside a reaction vessel, LMU researchers watch chemical reactions at work. Their results will improve the manufacture of the next generation of energy materials.
Shooting a movie in the lab requires special equipment. Especially when the actors are molecules – invisible to the naked eye – reacting with each other. “Imagine trying to film tiny lava flows during a volcanic eruption. Your smartphone camera wouldn’t be up to the job. First, you’d need to develop a special method to make the action you want to capture visible,” says Prof. Emiliano Cortés, Professor of Experimental Physics and Energy Conversion at LMU, also member of the e-conversion Cluster of Excellence.
But the effort is worth it – particularly when the product of the reaction is a promising energy material: so-called covalent organic frameworks (COFs). Still quite young, this material class has great potential for applications in battery technology and the manufacture of hydrogen. But despite 20 years of intensive research, scientists have been unable to fully elucidate what actually happens during the synthesis of COFs. As such, materials are often developed by trial and error.
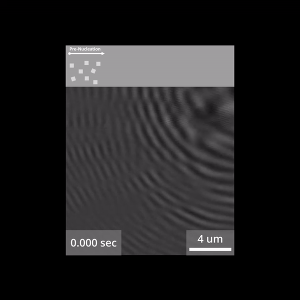
© Nano Energy Group / LMU
This has also been the case for COFs where several molecular components have to find the correct place during synthesis. Only then does the desired porous framework form over large areas. “Finding out why synthesis only works under certain conditions and not under others has intrigued me since my master’s days. Our approach in this project was to use the tools of physics to support chemists in their work. We wanted to shed more light on the complex synthesis processes and thus optimize them,” explains Christoph Gruber, who is researching this topic in Cortés’s team as part of his doctoral dissertation. To this end, the two scientists turned to the research group of LMU chemist Prof. Dana Medina, who is specialized in the synthesis of COFs, to establish a collaboration.
For the film shoot with the molecular stars, Gruber used a special microscope. With this tool, the team managed to follow the formation mechanism of the COFs at the nano level. The LMU researchers recently published their groundbreaking results in the journal Nature, accompanied by a video showing the processes that occur during synthesis in real time.
Synthesis of the molecular frameworks demands one thing above all: precise control of the reaction and self-assembly of the molecular building blocks present. “Only when you have this control it is probable to obtain a highly crystalline structure with an extensive order and, ultimately, the desired functionality,” says Medina. “However, our knowledge particularly of the early stages of nucleation and growth is full of gaps. And this has thwarted the development of effective synthesis protocols. We therefore were extremely intrigued to visualize the reaction as it unfolds and set the focus on the earliest stages when the mixed molecular components are starting to react.”
This is precisely where Gruber started with his investigations, choosing what would seem at first glance to be an unconventional method to cast light on the opening scene of COF formation: iSCAT microscopy. The abbreviation stands for interferometric scattering, and biophysicists often use this technology to investigate things like the interaction of proteins. “The measurement principle is based on the fact that even the tiniest of particles, made up of just a few molecules, scatter incident light. If these scattered light waves overlap, we get interference – just like water waves in a pool. That is to say, we get larger and smaller waves depending on how the waves overlap. We record these light patterns with a high-resolution camera and, with subsequent image processing, we obtain pictures that reveal, for example, nano-scale COF particles,” explains Gruber. And here’s the kicker: the iSCAT method is suitable for capturing dynamic processes and thus for real-time measurements. This allows the researchers to watch the synthesis live, as it were.
Imagine trying to film tiny lava flows during a volcanic eruption.PROF. EMILIANO CORTÉS
Immediately after the reaction started, the researchers were surprised to observe the presence of tiny structures in the transparent reaction medium. “The images showed us that nanometer-scale droplets can play an essential role in the synthesis. Although they are extremely small, they control the entire kinetics at the beginning of the reaction,” says Gruber. “Nothing was known about their existence before now, but for the formation of the COFs we studied, the nano-droplets turned out to be extremely important. If they are absent, the whole reaction happens too quickly and the desired order is lost.”
Using the iSCAT method, the LMU team managed to record a film showing the formation of the molecular frameworks from the beginning – with a sensitivity of just a few nanometers. “Existing techniques couldn’t capture the start of the reaction, with these nano-scale and millisecond-long processes, in real time,” says Cortés. “Through our research, we’ve now managed to close this gap in our knowledge. At the same time, we’re getting a holistic picture of the early stages of the reaction and the progressive formation of the COFs.”
Furthermore, the researchers used the film clip and the resulting analyses to design an energy-efficient synthesis concept. “Building on our results, we discovered how to rationally design the reaction conditions,” explains Medina. “By adding normal table salt, for example, we were able to massively reduce the temperature, such that the molecular frameworks form at room temperature as opposed to 120 degrees Celsius.”
The researchers are convinced their results will transform how we think about the synthesis of the over 300 different COFs and could therefore drive forward advances in industrial COF production. Moreover, the results could have far-reaching effects on the synthesis of other materials and on chemical reactions that have not yet been observed in real time. The LMU researchers are excited about shooting new films with molecules in the starring role.
Christoph G. Gruber, Laura Frey, Roman Guntermann, Dana D. Medina, Emiliano Cortés: Early stages of covalent organic framework formation imaged in operando. Nature, 2024.
2 May 2024
LMU researchers have deciphered the complex interplay of various enzymes around the innate immune receptor toll-like receptor 7 (TLR7), which plays an important role in defending our bodies against viruses.

The researchers used a wide range of technologies to track down the function of TLR7. | © jan Greune
Toll-like receptor 7 (TLR7), located in the dendritic cells of our immune system, plays a crucial role in our natural defense against viruses. TLR7 recognizes single-stranded viral and other foreign RNA and activates the release of inflammatory mediators. Dysfunctions of this receptor also play a key role in autoimmune diseases, making it all the more important to understand, and ideally modulate, the exact activation mechanism of TLR7.
Researchers led by Professor Veit Hornung and Marleen Bérouti from the Gene Center Munich and the Department of Biochemistry at LMU have now managed to gain deeper insights into the complex activation mechanism. It was known from earlier studies that complex RNA molecules have to be cut up first so that the receptor is able to recognize them. Using a wide range of technologies from cell biology to cryogenic electron microscopy, the LMU researchers have revealed how single-stranded foreign RNA is processed to be detected by TLR7. Their work has been published in the journal Immunity.
In the course of evolution, the immune system has specialized in recognizing pathogens from their genetic material. For example, the innate immune receptor TLR7 is stimulated by viral RNA. We can picture viral RNAs as long threads of molecules, which are much too large to be recognized as ligands for TLR7. This is where nucleases come in – molecular cutting tools that chop the ‘RNA thread’ into small pieces. Endonucleases cut the RNA molecules through the middle like scissors, while exonucleases cleave the thread from one end to the other. This process generates various RNA snippets, which can now bind to two different pockets of the TLR7 receptor. Only once both binding pockets of the receptor are occupied by these RNA pieces a signaling cascade set in motion, which activates the cell and triggers a state of alarm.
The researchers discovered that RNA recognition by TLR7 requires the activity of the endonuclease RNase T2 operating in conjunction with the exonucleases PLD3 and PLD4 (phospholipase D3 and D4). “Although it was known that these enzymes can degrade RNAs,” says Hornung, “we have now demonstrated that they interact and thereby activate TLR7.”
The researchers also found that the PLD exonucleases have a dual role within immune cells. In the case of TLR7, they have a pro-inflammatory effect, whereas in the case of another TLR receptor, TLR9, they have an anti-inflammatory effect. “This dual role of PLD exonucleases points to a finely coordinated balance for controlling appropriate immune responses,” explains Bérouti. “The simultaneous promotion and inhibition of inflammation by these enzymes could serve as an important protective mechanism for preventing dysfunctions arising in the system.” What role other enzymes could have on this signaling pathway and whether the molecules involved are suitable as target structures for therapies are subjects for further investigations.
Marleen Bérouti et al.: Lysosomal endonuclease RNase T2 and PLD exonucleases cooperatively generate RNA ligands for TLR7 activation. Immunity 2024
25 Apr 2024
LMU researchers decipher how an enzyme modifies the genetic material in the cell nucleus.
Inside the cell nucleus, the DNA molecule is found in a densely packed DNA-protein complex known as chromatin. To this end, the DNA is wrapped around a core of histone proteins and densely packed to form nucleosomes. The structure of the nucleosomes determines which genes are accessible and active and therefore plays an important role in gene regulation. To respond to metabolic signals, changed environmental conditions, and developmental processes, the nucleosomes must undergo repeated dynamic modifications with the aid of enzymes. A team led by Professor Johannes Stigler from LMU’s Gene Center Munich in collaboration with Felix Müller-Planitz (TU Dresden) has now carried out studies to investigate how a tiny chromatin modifying enzyme called ISWI remains mobile despite the densely packed material in the cell nucleus and is able to effectively rearrange nucleosomes.
The researchers were able to show that the enzyme consumes ATP – the energy currency of the cell – not only for its enzymatic activity, but also to navigate through the cell nucleus and to prevent the chromatin from becoming too rigid. “The movement of ISWI through the space densely packed with chromatin is powered by ATP. As it progresses, it keeps docking alternately with different binding sites on the nucleosomes. We compare this movement with that of a monkey swinging from branch to branch,” says Stigler. According to the researchers, the deciphering of these processes could yield insights into how defects contribute to diseases and could even open up new therapeutic avenues.
Petra Vizjak, Dieter Kamp, Nicola Hepp, Alessandro Scacchetti, Mariano Gonzalez Pisfil, Joseph Bartho, Mario Halic, Peter B. Becker, Michaela Smolle, Johannes Stigler, Felix Mueller-Planitz: ISWI catalyzes nucleosome sliding in condensed nucleosome arrays. Nature Structural & Molecular Biology 2024.
8 May 2024
Structural biologist Roland Beckmann renders the complex structures of ribosomes visible at the atomic level. In our interview, he talks about his research into the cellular protein factories.
Roland Beckmann, Professor of Biochemistry at LMU’s Gene Center Munich, investigates fundamental processes of life that occur in every cell. The structural biologist is a specialist in cryogenic electron microscopy, which allows the complex architecture of biomolecules to be made visible at the atomic level. He is particularly interested in the structure and workings of ribosomes, the protein factories of the cell. Together with Ed Hurt from Heidelberg University, Beckmann has summarized how ribosomes are assembled in the cells of higher organisms in two “snapshot” articles, which have appeared in the renowned journal, Cell. We spoke to Professor Beckmann about his research.
What occasioned the snapshots?
Roland Beckmann: We’ve reached a point where we – my team and Ed Hurt’s working together – have a very good overall understanding of the processes of ribosome biogenesis in eukaryotic cells – that is to say, the assembly of these complicated molecular machines. We’ve clarified many structures of intermediate stages and can now visualize the majority of this process, from the beginning in the nucleolus – a spherical area inside the cell nucleus – to the nucleus, to the transport from the nucleus into the cytoplasm. In view of this completeness, it made sense to publish these snapshots now and provide an overview.
Why are ribosomes important?
Ribosomes are one of the most important molecular machines we have. During the process known as translation, ribosomes link amino acids together according to the genetic information encoded in RNA to form proteins. And proteins of course are vital for pretty much all functions in the cell.
But the ribosome does much more besides: We like to compare it to a smartphone, with which you can make calls but also do a thousand other things. Ribosomes play an important role in quality control, for example, protecting the cell against defective and harmful translation products and keeping the amount of RNA molecules in equilibrium.
This quality control is our second major research field after ribosome biogenesis. We discovered, for instance, that ribosomes react sensitively to stress, whereupon they stall and bump into each other during the translation process. They have traffic accidents, so to speak. And the cell is able to recognize this and trigger protective mechanisms. These collisions have proven to be a general principle that holds from bacterial to human cells. Very broadly, we can say that the ribosome is used as a sort of integrator for perceiving environmental influences, which can then be read by the cell to trigger the necessary reactions.
But the ribosome does much more besides: We like to compare it to a smartphone, with which you can make calls but also do a thousand other things.ROLAND BECKMANN
The snapshots summarize the results of ten years of research into ribosome biogenesis. What were the important milestones on this journey?
The ten years refer to the collaboration between my research group and Ed Hurt’s; I myself have been researching this topic for a good deal longer. The ribosome consists of a large and a small subunit, and our first major milestone, I would say, was when we elucidated the structure of a precursor of the small subunit, which had only been known biochemically. This so-called processome is interesting because it is extremely large. Indeed, it is larger than the full-grown ribosome and has a shape that allows the small subunit to be folded into it.
Subsequently, we found more and more intermediates and discovered that the small subunit peels off the processome like an onion, with one component after another being shed. And we’re also confident that a certain state of the small subunit is necessary for the peeling-off signal to be given.
And the large subunit?
With the large subunit, it was a similar story. At first, it was not at all clear how it starts to assemble. We identified very early intermediates and showed that first an exoskeleton is formed, which is filled up rather like you would fill a bowl. That was entirely unexpected.
In addition, we discovered a puzzling phenomenon whereby a certain RNA is incorporated into the large subunit the wrong way round. This RNA forms a sort of arch, which is initially turned round 180 degrees. Later, the position is corrected by means of rather complicated machinery. This backward integration, it turns out, has been preserved from bacteria to humans and is designed to prevent incorrect folding in the course of assembly.
Other results have shown that there is clearly a general principle by which the RNA is taken out of play, as it were, through the binding of certain factors, so that it does not get in the way of other areas when they are being folded. The final conformation is only adopted with the progressive completion of the ribosome.

Structural biologist Roland Beckmann | © LMU
What makes working with ribosomes so challenging?
First of all, the intermediate products that arise during assembly are highly complex. These assembly intermediates consist of large RNAs and hundreds of proteins. It’s thought that human cells need 300 or more factors to make these complexes. It’s not so easy here to isolate individual substages, nor can you manufacture hundreds of factors and reproduce the assembly in vitro – particularly since assembly begins at the instant in which the ribosomal RNA is transcribed.
Moreover, we can’t really get at the very early intermediate products. This is partly because they are so unstructured that it’s scarcely possible to meaningfully analyze them, but also because it’s so hard to isolate them. Many intermediates are in the nucleolus, and that is one of the classic organelles without a membrane. The nucleolus represents a condensate with unique properties of its own, and it’s difficult to get at the corresponding molecules to study them.
On the other hand, ribosomes are otherwise very good objects for analysis using cryogenic electron microscopy, where the motto is: the bigger, the better.
It was important for us to get a handle on the model systems by developing certain markersROLAND BECKMANN
Has technical progress played an important role over the past decade?
Absolutely. It was important for us to get a handle on the model systems by developing certain markers. Because we cannot assemble the intermediate stages, we have to isolate them using specific affinity tags. The problem is that the tags can be bound for rather a long time during the manufacture of the ribosome and migrate from the nucleolus to the cytoplasm along with various intermediates. As a result, you get a menagerie of intermediate products during the cleaning, which cannot be adequately sorted.
Ed Hurt invented beautiful systems that permit the use of two different markers. This is referred to as a split tag. It allows us, for example, to first tag one factor, which is bound to the intermediates from the nucleolus all the way into the nucleus, and then tag a second factor, which might be bound only from the end phase in the nucleus to the cytoplasm. In this way, we can isolate smaller subgroups. That was absolutely key to all the things we’ve done.
A second important aspect is that we started at a relatively early stage to work not only with yeast as a model system, but also with a thermophilic fungus. These fungi live in dung heaps, where temperatures can reach up to 50-60 degrees. Consequently, their proteins are adapted to heat and are much more stable and better to handle at room temperature than their yeast counterparts.
The best resolutions approach one Ångström. And naturally this makes a big difference.ROLAND BECKMANN
Have there also been advances in microscopy?
Yes, that was the second thing that was absolutely decisive. There have been huge technical advances in cryogenic electron microscopy. The first detectors had scintillators that generated a light pulse with each electron, which was then seen by the detector. These detectors were replaced by direct electron detectors, which no longer need a scintillator and can actually perceive each individual electron with pixel resolution.
This improved the quality to such a degree that molecular resolution – that is to say, three Ångström or better – is routinely achieved, which was previously unobtainable. The best resolutions approach one Ångström. And naturally this makes a massive difference. In addition, there have been new developments in software for the reconstruction and classification of particles.
Can defective ribosomes trigger diseases?
In principle, yes. Ribosomopathies do in fact exist. These diseases are based in most instances on mutations in ribosomal proteins or in assembly factors, which cause ribosomes to be formed more slowly or incorrectly. Astonishingly, most of these diseases affect blood formation, leading to anemias or leukemias.
But in principle, interventions are certainly possible, if you have understood the mechanisms.ROLAND BECKMANN
Can your research help pave the way for new therapeutic approaches?
One of the reasons we started investigating this subject was the connection between ribosomopathies and defective assembly reactions. If we understand the mechanisms, we surmised, then it might lead to something. However, this is a difficult step to accomplish, because, as it transpired, a pathological phenotype is caused by the ribosome quantity being wrong. Correcting a deficit is rather tricky in this complex context. But in principle, interventions are certainly possible, if you have understood the mechanisms.
The faulty ribosomes are dismantled, leaving the cells without enough ribosomes. This presumably means that certain RNAs that do not occur frequently in the cell are no longer converted into proteins. Consequently, a different set of proteins is made than would be the case with a normal quantity of ribosomes. There are very many different mutations we are now aware of. This leads to an imbalance in translation, which we don’t yet understand well.
This is one question we’re studying. We want to investigate, for example, where these sick ribosomes end up. Are they all dismantled, or do some of them wind up in the translation cycle after all, where they might cause stress?
What questions remain to be resolved?
There are many open questions relating to ribosome biogenesis. A major problem is that although our structural investigations produce beautiful galleries, they are rather static and often we don’t know exactly how one state leads to another. Which signals are needed for the process to advance? This is quite difficult to address, partly because in-vitro investigations are so rarely successful. In fact, you can only really observe these things in the cell. In bacteria, we can reconstitute the process isolated in the test tube to a certain degree, but we haven’t accomplished this in eukaryotic cells yet. It would be interesting to really understand the transition that takes place in ribosome biogenesis and its incorporation into all possible signaling pathways.
And when it comes to translation, there’s also a lot we don’t yet understand. For example, how does the recognition process actually work when ribosomes collide. How do they realize that they have bumped into each other? How are the ensuing signaling pathways generated at the molecular level? And how are these signals related to each other? As you can see, there are many outstanding questions. Our work is far from done.
24 Jun 2024
The Gene Center Munich stands for interdisciplinary top-level research. As the center turns 40, we interview its director Karl-Peter Hopfner, who recalls the most important stages in its history and talks about the challenges to come.

The Gene Center moved to its current building in Großhadern in 1994. | © LMU/Jan Greune
Four decades of top-level research in the life sciences: the Gene Center Munich celebrates its 40th birthday on 24 June. Founded in 1984 as a joint institute of LMU and the Max Planck Institute of Biochemistry, it combines interdisciplinary research with the fostering of early-career scientists in key areas of the life sciences and shapes the life sciences in Munich and further afield. Professor Karl-Peter Hopfner has headed the center since 2015.
What’s special about the Gene Center?
Karl-Peter Hopfner: On top of its mission to strengthen molecular biology in the German research landscape, the Gene Center Munich has had two other main features since its foundation. One is its strong interdisciplinarity, which we encapsulate in the slogan, “Beyond Disciplines.” Thanks to its special organizational structure, the Gene Center has the capability – outside the classical faculties – to create links between faculties and bring expertise from different fields together in one building.
The second feature is the center’s leading role in promoting early academic independence. Just as important as interdisciplinarity, this feature has been there from the outset. The fostering of early-career research groups began with the establishment of the institution and was further developed in the so-called tenure-track model, which enables researchers to be appointed to a professorship directly after the postdoc phase – bypassing the traditional habilitation. This appointment is then made permanent following an evaluation.
We have maintained and expanded these two emphases over the past four decades and they will remain the lodestars for the Gene Center in the university landscape of the future.
What was the goal when the Gene Center was founded?
Hopfner: At the start of the 1980s, when molecular biology was an emerging discipline, Ernst-Ludwig Winnacker and colleagues realized that it would not only open up groundbreaking new research possibilities within the life sciences, but also pave the way for exciting medical and industrial applications. Our center is one of four centers that were founded back then as part of a nationwide gene center program designed to keep and establish this fledgling technology in Germany and recruit young researchers for this purpose. These centers were called “Gene Centers” at that time and, in our case, the name stuck. The founding name of the institute was the Laboratory for Molecular Biology.
It was established in 1984 under founding director Ernst-Ludwig Winnacker as a joint institution of LMU and the Max Planck Institute of Biochemistry, in the premises of which the Gene Center was housed for its first ten years, before our building on Campus Grosshadern was inaugurated.

© LMU/Jan Greune
What are your main research areas?
Hopfner: We have three mainstays: One is basic research in the life sciences – that is, the question as to how cells and the molecules in cells work.
The second mainstay has developed over the past ten to fifteen years. This is the domain of molecular systems biology, in which we investigate how molecules cooperate to form living systems. Molecular systems biology encompasses fields such as gene regulation, the immune system, cell-cell communication, and cell differentiation.
In the first domain, then, we have a highly focused view on molecules as soloists, so to speak, whereas in the second domain we take a broader view on the orchestra of many molecules.
The third mainstay is clinical translation, which involves groups from the clinical sciences, from LMU University Hospital, and from the Faculty of Veterinary Medicine. The focus here is on taking the findings from research and developing them with an eye to therapeutic applications.
Looking back on the history of the Gene Center, which events shaped its development?
Hopfner: After the foundation and the first phase in the Max Planck Institute of Biochemistry, the construction of our building in 1994 was certainly a milestone – and a significant event in the development of Campus Grosshadern as well. Apart from LMU University Hospital Grosshadern and the Max Planck Institute, this was all just fields and farms. Then came the Gene Center, followed by the chemistry buildings, the biology buildings, and the Biomedical Center. All that developed around the Gene Center to an extent. And now we have this wonderful campus, which is one of a kind in the life sciences in Germany. The Gene Center played a pivotal role in shaping this evolution.
Another important development was the idea of Rudi Grosschedel, who headed the Gene Center from 1998 to 2003, to establish the tenure-track model, inspired by his experience in the Anglo-American academic sphere. I think that was a brilliant strategic move, including on the part of the university and the faculty, who all pulled together to make it a reality. It has really helped us be competitive in the recruitment of excellent researchers from all over the world over the past 20 years.
And now we have this wonderful campus, which is one of a kind in the life sciences in Germany. The Gene Center played a pivotal role in shaping this evolution.KARL-PETER HOPFNER
How does the story continue?
Hopfner: Further milestones are related to the excellence initiative. In this context, Patrick Cramer in particular hugely expanded top-level research at the Gene Center from 2004 to 2013. Together with colleagues from the chemistry faculty, we founded the first cluster of excellence in the form of CIPSM (Center for Integrated Protein Science Munich). This was an important step for the Gene Center, and later we had a Collaborative Research Center funded by the German Research Foundation, where we demonstrated that we could pursue excellent joint research here.
In the next round of the excellence initiative, Ulrike Gaul, who came here as a Humboldt Professor in 2009, worked very hard with Patrick Cramer to establish systems biology at the Gene Center and organized the QBM graduate school, which bridged biochemistry and physics.

The BioSysM (Research Center for Molecular Biosystems) was inaugurated in 2016. | © LMU
And then BioSysM came along later?
Hopfner: Yes, we grew so much over time that it was getting cramped here. And so Patrick Cramer led an initiative together with Ulrike Gaul to apply for an extension – what would become BioSysM (Research Center for Molecular Biosystems) – which was inaugurated in 2016. Through the establishment of BioSysM, we gained a second axis, as it were, which principally covers the field of molecular systems biology. Meanwhile, structural biology and translational research are mainly located in the original Gene Center. The art of course is to synergistically expand and bring together these different research strands.
Is it accurate to describe the Gene Center as an incubator for interdisciplinary collaboration?
Hopfner: Indeed. One strength of the Gene Center is that the whole is greater than the sum of the individual parts. Naturally, this only works if there are synergies and people work well together. We have a relatively large number of publications that were produced by multiple groups. When you appoint people and see that three years later, they publish a joint paper that is celebrated by peers, what more do you want?
Interdisciplinarity was always very important to me, and it’s a quality that’s baked into the Gene Center. By virtue of the groups here with members from four faculties, you can say we live the interfaculty creed, and this gives rise to a plethora of collaborations. Moreover, it’s something I’d like to further expand.
One strength of the Gene Center is that the whole is greater than the sum of the individual parts.KARL-PETER HOPFNER
Are there synergies in teaching as well?
Hopfner: Yes, we’re very active in this regard. Together with the Department of Chemistry, we have a bachelor’s degree course built on a so-called Y-model. The students start out together and branch off later, either in the direction of chemistry or of biochemistry. This is a unique course in the German research landscape, one that has many USPs and is highly attractive. We’re currently thinking about expanding this portfolio with somewhat more specialized bachelor’s degree courses. We also want to forge new paths here – for example, by connecting systems biology and biochemistry.
In addition, we have an international master’s degree course in English, where 40-50 percent of students come from abroad. We also help our students in the master’s program get to know the world – say, by doing their thesis abroad. This is a great and relatively straightforward opportunity to experience research in a different country.

© LMU/Jan Greune
Can you name examples of innovations that were developed at the Gene Center?
Hopfner: My favorite example comes from the Klaus Conzelmann group, which worked on viral systems. These included rabies viruses, which attack nerve cells. The group developed a system whereby the viruses, by means of a fluorescent dye, stain the cells they attack – along with all the cells directly connected to them. In this way, scientists can visualize how these cells interact in the brain. This was a stunning achievement and the system is used by neurobiologists worldwide to map how nerves are configured in the brain.
Another brilliant innovation from the Gene Center – and one that may not be on people’s radar – was developed by Johannes Söding and staff and actually comes from the field of computational biology. Söding’s team developed a method for comparing amino acid sequences and searching for weak homologies – that is to say, distant relationships – making it possible to construct very large relationship maps across all known life forms. This technology was instrumental in the development of the AI program AlphaFold2. These types of AI models, which can not only predict the structure of proteins, but also generatively design them, are partly based on analyses made possible by the technology created by Söding and his team.
And then of course we’ve done a lot of spectacular things in basic research – in areas such as transcription machinery, ribosome biology, the innate immune system, DNA repair, mitochondrial stress, and chromatin. Now, these are not classical innovations in the form of patents, but they are science that will make its way into the next generation of biochemistry textbooks. These are our two spheres – basic research and applied research.
Many researchers at the Gene Center were way ahead of their time.KARL-PETER HOPFNER
No doubt, the technological possibilities have also changed hugely over the past 40 years …
Hopfner: I don’t need to go back 40 years for that – even over the last four years, the world of research has dramatically changed. Rapid technological advances will undoubtedly transform our work in the future, opening up brand new possibilities. I must say that many researchers at the Gene Center were way ahead of their time. For example, we had scientists working on gene therapies and protein design and protein folding. These were wonderful projects, which were not really effective at that time in the late 90s because certain technological advances had not yet been made. The scientists couldn’t work with the accuracy of structure prediction required for protein design, for example, or the methods of gene therapy weren’t yet accurate or efficient enough.
But these fields of technology and development are coming back strong – for example, as a result of CRISPR/Cas9. Gene therapy is currently one of the fastest developing therapeutic approaches alongside mRNA technology.
And protein design has also progressed thanks to generative AI. The AI model AlphaFold and AI developments in protein design will open up whole new worlds, including in the therapeutic sphere and in biotechnology. This rapid progress is also a big challenge for an institute like ours, because naturally you have to plan ahead a bit. Our strategy is to set ourselves up on a relatively broad technological basis, so that we can pursue as many directions as possible, and embed the whole in a research topic that binds us together.
Our strategy is to set ourselves up on a relatively broad technological basis, so that we can pursue as many directions as possible, and embed the whole in a research topic that binds us together.KARL-PETER HOPFNER

Professor Karl-Peter Hopfner heads the Gene Center Munich since 2015. | © jan greune
Which topic will provide your collective superstructure in future?
Hopfner: Nucleic acid biology, which has seen huge advances in recent times. First of all, the study of nucleic acids – that is to say, RNAs and DNAs – looks at fundamental processes in the cell. This includes how nucleic acids work in the cell, how proteins bind to them, and how DNA damage and diseases arise. Secondly, nucleic acids, particularly in the form of RNA, have become a therapeutic entity and are currently at the heart of much translational research. That’s why they’re an important topic here at the Gene Center.
What is your vision for the future of the Gene Center?
Hopfner: To build on our motto “Science Beyond Disciplines.” We want to introduce more AI so that we can participate in fields such as generative AI and design. If we achieve this and are attractive to talented people from all over the world thanks to our flat hierarchies and interdisciplinarity, then we will be in an excellent position over the years to come as regards pursuing top-level research
9 Apr 2024
Andreas Kornath, born in Bergkamen in 1965, passed away on March 5, 2024 at the age of 58.

Professor Andreas Kornath passed away on March 5, 2024.
After graduating from high school in 1985, Kornath began studying chemistry at the University of Dortmund, which he completed in 1989. He continued his academic career with a diploma thesis under the supervision of Professor Rolf Minkwitz on the chemistry of mercaptosulphonium salts, which he successfully completed in September 1990.
In 1993, he received his doctorate with a thesis on the chemistry of chalcogen and picogenium salts and triphenylsilysulphanes under Professor Minkwitz in Dortmund.
This was followed by a post-doctorate at the University of Alabama in Tuscaloosa, USA. Kornath then returned to Germany and worked as a research assistant at the University of Dortmund from January 1995 to September 2000. During this time, he also spent three months at the University of California in Los Angeles.
Andreas Kornath habilitated in Dortmund in 2000 with a thesis on the structural elucidation of ligand-free metal cluster reactivity of bare anions. Kornath subsequently worked there as a lecturer and later as a senior research associate in the Department of Inorganic Chemistry. Between April 2005 and July 2006, he held professorships in inorganic chemistry at the universities of Rostock and Munich. From August 2006, he worked as a university lecturer in Dortmund before taking up a professorship in inorganic chemistry at LMU in April 2007, which he held until the end of his life.
Kornath's research focused on extremely strong acids, also known as superacids. He was particularly interested in the associated phenomena, which are important not only on Earth but also in interstellar space.
Acid strength plays a decisive role in numerous chemical reactions in biological and technical processes. Superacids reach the highest known acidities and therefore offer enormous potential for the investigation of highly reactive intermediates as well as for applications in technical processes.
Andreas Kornath's scientific work, characterized by his commitment to research and teaching, has significantly enriched the world of inorganic chemistry. His legacy will endure through his numerous publications and the generations of chemists he trained and inspired.
10 Apr 2024
LMU researchers decode repair mechanism during transcription of genetic information
© @ LMU / Jan Greune
Cockayne syndrome is a severe autosomal recessive disorder caused by defective DNA repair mechanisms. People with the disease have much reduced life expectancy and suffer from facial deformities; growth failure; neurological, cognitive, and sensory impairments; bone, joint, and muscle deformities; kidney problems; and premature aging. Like xeroderma pigmentosum (XP), Cockayne syndrome (CS) is a disease where elements of nucleotide excision repair (NER) do not work properly. The purpose of this repair mechanism is to remove DNA damage caused by ultraviolet (UV) light, chemicals, and various other environmental factors.
Researchers from the group of biochemist Professor Julian Stingele from LMU’s Gene Center Munich have now uncovered important details about the role of the CSB/ERCC6 and CSA/ERCC8 genes involved in Cockayne syndrome. These genes encode two enzymes associated with DNA repair. The results of their work have been published in the journal Nature Cell Biology. “Our data point to a new, previously unknown function of these two genes and their gene products in the repair of covalent DNA-protein interactions in the course of transcription,” reports Stingele, referring to the cytotoxic, biologically undesirable crosslinking of proteins to DNA.
In collaboration with researchers from the University of Cambridge, the scientists demonstrated that DNA-protein crosslinks present a physical obstacle to further transcription. Arresting transcription brings CS proteins to the blockade sites. “Our results indicate that CSB and CSA then initiate the transcription-coupled repair of the toxic DNA-protein crosslinks,” says Stingele. “This previously unrecognized cellular function of CS proteins leads to the marking of the DNA damage – and thence to its enzymatic breakdown.”
The study also revealed that this newly discovered function of CS proteins works independently of classic TC-NER (transcription-coupled nucleotide excision repair) enzymes, which are deployed, among other things, for repairing DNA damage caused by UV light – and the absence of which leads to xeroderma pigmentosum. “The fact that CS proteins have additional functions is noteworthy. This discovery could help to explain the pathological differences between xeroderma pigmentosum and Cockayne syndrome,” says Stingele. CS is a more severe and more multifaceted disorder than XP, with complex and incompletely understood causes. As their next step, Stingele’s research group plans to decode the exact process of CS-protein-mediated repair.
Christopher J. Carnie et al.: Transcription-coupled repair of DNA-protein crosslinks depends on CSA and CSB. Nature Cell Biology 2024
13 Mar 2024
Following a decision by the Faculty Library Commission, LMU's Chemistry and Pharmacy Library has joined the Open Access (OA) transformation agreement for Germany negotiated between TIB Hannover and Royal Society of Chemistry (RSC) in 2024.

Here are the key points of this new RSC Platinum model:
Further information and the current list of titles on which the agreement is based can be found here.
If you have any questions or problems in connection with the RSC agreement, please contact your subject librarian:
Dr. Andreas Will
andreas.will@ub.uni-muenchen.de
+49 89 2180 77065
10 Jan 2024
On December 8, 2023, a group of students visited our faculty as part of LMU's University Day.

The University Day is an LMU event in which a group of selected high school students from grammar schools in Upper Bavaria visit the university's various faculties over the course of a semester. On December 8, 2023, the group visited the Departments of Chemistry and Biochemistry.
The pupils learned about our combined chemistry/biochemistry degree program and our teaching degree program, took part in a lecture and enthusiastically carried out their own experiments in various laboratories.
14 Feb 2024
Researchers at LMU have developed an innovative method to simultaneously track rapid dynamic processes of multiple molecules at the molecular scale.

Fiona Cole and Jonas Zähringer, joint lead authors of the paper, calibrate a fluorescence microscope. © LMU
Processes within our bodies are characterized by the interplay of various biomolecules such as proteins and DNA. These processes occur on a scale often within a range of just a few nanometers. Consequently, they cannot be observed with fluorescence microscopy, which has a resolution limit of about 200 nanometers due to diffraction. When two dyes marking positions of biomolecules are closer than this optical limit, their fluorescence cannot be distinguished under the microscope. As this fluorescence is used for localizing them, accurately determining their positions becomes impossible.
This resolution limit has traditionally been overcome in super-resolution microscopy methods by making the dyes blink and turning their fluorescence on and off. This temporally separates their fluorescence, making it distinguishable and enabling localizations below the classical resolution limit. However, for applications involving the study of rapid dynamic processes, this trick has a significant drawback: blinking prevents the simultaneous localization of multiple dyes. This significantly decreases the temporal resolution when investigating dynamic processes involving multiple biomolecules.
Under the leadership of LMU chemist Professor Philip Tinnefeld and in cooperation with Professor Fernando Stefani (Buenos Aires), researchers at LMU have now developed pMINFLUX multiplexing, an elegant approach to address this problem. The team recently published a paper on their method in the journal Nature Photonics. MINFLUX is a super-resolution microscopy method, enabling localizations with precisions of just one nanometer. In contrast to conventional MINFLUX, pMINFLUX registers the time difference between the excitation of dyes with a laser pulse and the subsequent fluorescence with sub-nanosecond resolution. In addition to localizing the dyes, this provides insights into another fundamental property of their fluorescence: their fluorescence lifetimes. This describes how long, on average, it takes for a dye molecule to fluoresce after it is excited.
“The fluorescence lifetime depends on the dye used,” explains Fiona Cole, co-first author of the publication. “We exploited differences in fluorescence lifetimes when using different dyes to assign the fluorescent photons to the dye that emitted without the need for blinking and the resulting temporal separation.” For this purpose, the researchers adapted the localization algorithm and included a multiexponential fit model to achieve the required separation. “This allowed us to determine the position of multiple dyes simultaneously and investigate rapid dynamic processes between multiple molecules with nanometer precision,” adds Jonas Zähringer, also co-first author. The researchers demonstrated their method by accurately tracking two DNA strands as they jumped between different positions on a DNA origami nanostructure, as well as by separating translational and rotational movements of a DNA origami nanostructure and by measuring the distance between antigen-binding sites of antibodies. “But this is just the beginning,” says Philip Tinnefeld. “I am certain that pMINFLUX multiplexing, with its high temporal and spatial resolution, will provide new insights into protein interactions and other biological phenomena in the future.”
INFO: Fiona Cole, Jonas Zähringer, Johann Bohlen, Tim Schröder, Florian Steiner, Martina Pfeiffer, Patrick Schüler, Fernando D. Stefani & Philip Tinnefeld: Super-resolved FRET and co-tracking in pMINFLUX. Nature Photonics 2024
5 Mar 2024
Using a so-called computational hyperreactor, LMU chemists have managed to calculate highly complex chemical reaction networks efficiently under realistic conditions.

Molecular reaction networks that reveal the various reaction possibilities of a system are of vital importance for chemistry and biochemistry. The molecular origins of life are a prime example of branched networks. We need only recall the famous “primordial soup” experiment (Miller-Urey experiment) of 1953 on the emergence of organic molecules in the Earth’s early atmosphere or consider the formation of molecules in interstellar space. Understanding, predicting, and ultimately calculating such reaction networks is a highly complex task. With the development of a so-called hyperreactor, the team led by Christian Ochsenfeld, Professor of Theoretical Chemistry at LMU, is pursuing this ambitious goal.
Using the tools of theoretical chemistry, it is possible to simulate such reaction networks. A key challenge is presented by the complexity of the potential energy hypersurfaces of chemical systems, with numerous minima and connecting saddle points. The latter constitute energy barriers which a chemical reaction must overcome. Simplifying matters, we can picture this potential energy hypersurface as a mountainous terrain, where various molecules or structures correspond to the valleys and the energy barriers to be overcome are represented by the mountains.
The difficulty posed by the energy barriers, which are often very high, were circumvented in early approaches by means of simulation under periodic contractions (pressure) and very harsh reaction conditions such as extremely high temperatures to induce chemical reactions. However, this has also led to unrealistic reactions and fragmentations. By contrast, the new hyperreactor from the Ochsenfeld group enables the study of complicated reaction networks under mild conditions. To this end, periodic contractions of the molecular system are coupled with the exploration of chemical space on smoothed potential energy surfaces.
Sample applications of the hyperreactor – such as the calculation of nonenzymatic DNA nucleoside synthesis, which is being experimentally researched in the group led by Oliver Trapp, Professor of Organic Chemistry at LMU, or the synthesis of glycinal, acetamide, and carbamic acid in interstellar ices at -263°C – are yielding initial insights into the new computational possibilities. Particularly when combined with new, almost 1,000 times faster quantum chemical methods, which were also developed in Ochsenfeld’s research group, the hyperreactor opens up new perspectives for the exploration of complex reaction networks and for research into a wide variety of chemical and biochemical synthesis pathways.
Alexandra Stan-Bernhardt, Liubov Glinkina, Andreas Hulm, Christian Ochsenfeld: Exploring Chemical Space Using Ab Initio Hyperreactor Dynamics; ACS Central Science 2024
1 Mar 2024
Researchers at LMU and Stanford University reveal how cells regenerate protein factories at endoplasmic reticulum.
The protein UFM1 acts as a molecular wedge, facilitating the cleavage of the Sec channel. © T. Becker, Nature 2024
The synthesis of proteins in the cell is a key process of life. By this means, the genetic code of the genome is translated into the amino acid sequence of proteins. The process is complex – and has been studied in detail for decades.
Protein biosynthesis is performed by special molecular machines, ribosomes, which consist of a large and small subunit. At the end of protein biosynthesis, these protein factories have to be broken up into their individual parts (recycled), so that they are ready for the next round of translation.
Now a team led by Professor Roland Beckmann, Dr. Thomas Becker, and Ivan Penchev from LMU’s Gene Center Munich, working in collaboration with researchers at Stanford University led by Professor Ron Kopito, have shown how the recycling of ribosomes at the so-called endoplasmic reticulum (ER) functions. In the process, they discovered the role of an enzyme, a special E3 ligase that joins a small protein modification called UFM1 to the large ribosomal subunit, as a key mechanism of recycling. An account of their investigations has been published in the prestigious journal Nature.
Ribosomes are usually found floating within the cytoplasm. “Here we know precisely how the recycling works,” says Becker. Sometimes, however, they are located at the endoplasmic reticulum, a continuous cell-wide membrane network.
Although many proteins originate in the cytosol, they subsequently have to be brought to other organelles, such as mitochondria, chloroplasts, and many more. If a protein is synthesized at the ER membrane, the whole translation machinery docks to the ER membrane. This is accomplished with the aid of a protein-conducting channel (SEC61), which is able to transport proteins across the membrane or insert them into the membrane during synthesis.
After completion of the translation, there is a further recycling step, which is specific to the ER membrane. Namely, the large subunit of the ribosome has to be detached again from the protein-conducting channel. Beckmann’s team has now demonstrated how this substep works: When the translation has finished, the E3 ligase recognizes the large subunit of the ribosomes. “It places – figuratively speaking – a small wedge, the protein UFM1, at the large subunit,” explains Becker. "This produces a stable complex out of the modified 60S subunit and the E3 ligase. Simultaneously, it causes the large subunit to detach from SEC61. This is a very important step to ensure that the large subunit is returned to the cytosol and available for the next round."
Paul A. DaRosa, Ivan Penchev, Samantha C. Gumbin, Francesco Scavone, Magda Wąchalska, Joao A. Paulo, Alban Ordureau, Joshua J. Peter, Yogesh Kulathu, J. Wade Harper, Thomas Becker, Roland Beckmann & Ron R. Kopito: UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER. Nature 2024