Gene regulation - Histone exchanger comes into focus
Most of the genomic DNA in the cell nuclei of all higher organisms is associated with so-called histone proteins, forming a complex known as chromatin. Chromatin is basically made up of subunits called “nucleosomes” – each consisting of a segment of DNA wound around a histone core like thread coiled on a spool. The nucleosomal DNA may be further compacted by interaction with certain types of non-histone proteins. The packing is modified by so-called chromatin remodeling complexes, which locally regulate the accessibility of the DNA by displacing nucleosomes along the double helix and/or replacing specific histones within the nucleosome core. These remodelers are essential for cell function and survival, since nucleosomes repress gene expression as well as regulating other processes such as the recognition and repair of DNA damage.
“Little is known about the structure and mode of action of chromatin remodelers, because these molecules are large, complex and highly flexible – all of which makes their architecture difficult to study using conventional methods,” says Professor Karl-Peter Hopfner. He and his research group at LMU’s Gene Center have already determined the structures of some of the smaller and relatively simple remodeling proteins. Now, in close collaboration with his LMU colleague Professor Roland Beckmann (Gene Center and Department of Biochemistry), Hopfner has achieved a breakthrough in the structural analysis of a more complicated, chromatin-associated, molecular machine. With the aid of a novel combination of structural and biochemical techniques, the LMU researchers have succeeded for the first time in determining the overall architecture of a large chromatin remodeler.
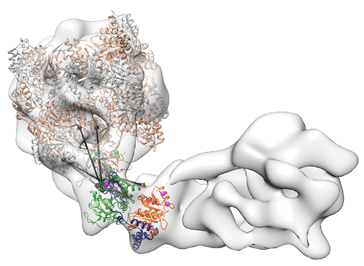
The multisubunit remodeling complex in question is called INO80, a remodeler and histone exchanger that is recruited to sites of DNA damage. Initial analysis by electron microscopy revealed that its structure is very different from those of the smaller remodelers investigated hitherto. INO80 has an elongated shape, is capable of adopting a flexed, embryo-like conformation, and lacks the kind of cleft, which is postulated to accommodate the nucleosome in the remodelers so far studied. “However, the resolution of the 3-D reconstructions derived from electron micrographs does not allow us to distinguish the individual subunits that make up the INO80 complex,” says Caroline Haas, joint first author of the new study.
“In order to get a clearer picture of the structure, we took advantage of a method which was originally developed in Professor Ruedi Aebersold’s laboratory at the ETH in Zürich, and was established in our group at the Gene Center by Franz Herzog,” says Hopfner. The method employs a bifunctional reagent that reacts with chemical groups that are a certain distance apart, thus linking proteins that are close together in the complex. The use of a mixture of isotopically labeled versions of the chemical permits the subsequent identification of the cross-linked sites by mass spectrometry. Based on the resulting patterns, a protein-protein interaction map can be assembled that reveals aspects of the topology of the complex. “With this method, one can – within limits – deduce where in the complex each subunit lies,” as Alessandro Tosi and Franz Herzog, also joint first authors of the new report, point out.
By combining results from electron microscopy and mass spectrometry, the researchers obtained insights into how the remodeler recognizes its nucleosomal substrate. They inferred that the complete complex is probably required for binding of the nucleosome, which is enveloped within the embryo-like flexure, displacing the “foot” of the complex. “Our structural model provides a basis for dissecting how INO80 mediates the exchange of histone variants in nucleosomes – and thus represents an important step toward a detailed understanding of the workings of this class of large and adaptable molecular machines,” Hopfner concludes.
The work was financed by the DFG in the context of Collaborative Research Center (SFB) 646, SFB/Transregio 5 and Graduate School (GRK) 1721, and supported by the Research Network for Molecular Biosystems (BioSysNet) and the Center for Integrated Protein Science Munich (CIPSM), a Cluster of Excellence.
Publication: Structure and subunit topology of the INO80 chromatin remodeler and its interaction with the nucleosome, Alessandro Tosi, Caroline Haas, Franz Herzog, Andrea Gilmozzi, Otto Berninghausen, Charlotte Ungewickell, Christian B. Gerhold, Kristina Lakomek, Ruedi Aebersold, Roland Beckmann and Karl-Peter Hopfner, Cell 2013